Generative Models¶
To get the conditional probability P(Y|X), generative models estimate the prior P(Y) and likelihood P(X|Y) from training data and use Bayes rule to calculate the posterior P(Y |X):
we have a notion of the underlying distribution of the data and we want to find the hidden parameters of that distribution. Some approaches are:
- Naive Bayes classifiers,
- Gaussian mixture models,
- variational autoencoders,
- generative adversarial networks and others.
Discriminative Models¶
discriminative models directly assume functional form for P(Y|X) and estimate parameters of P(Y|X) directly from training data. They are more suitable when we only want to find the boundary that separates the data into different classes.
- support vector machines
- logistic regression (LR),
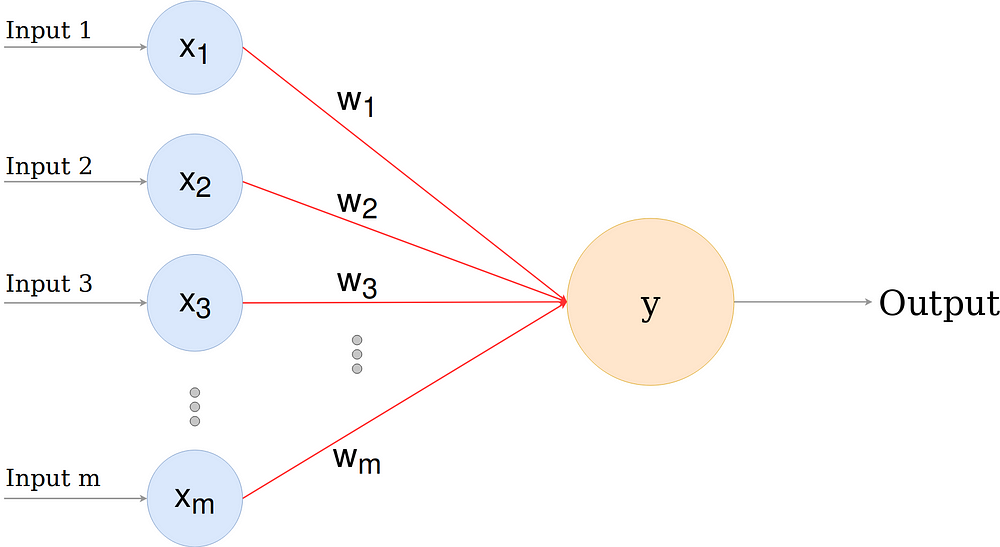
- perceptron